BugView
A Genome Browser for the Desktop

Introduction
‘BugView’ is a standalone genome browser for comparing the arrangement of genes on a pair of related genomes, although it can also be used to view individual genomes. It was written for comparative studies of bacterial genomes — hence the name — although it handles intron-containing eukaryotic chromosomes equally well. As a desktop application, rather than a web facility, ‘BugView’ responds quickly to adjustment of the visualization, the user has control over file input (GenBank format is used), and views of the genomes can easily be printed or saved.
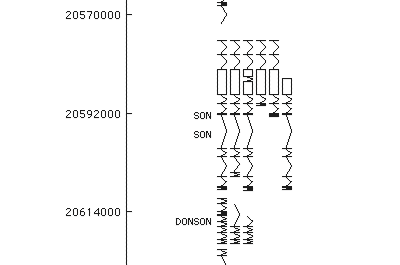
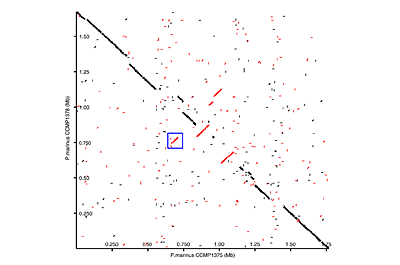
Click on the thumbnails below for screen-shots illustrating some of the visualizations that ‘BugView’ provides.
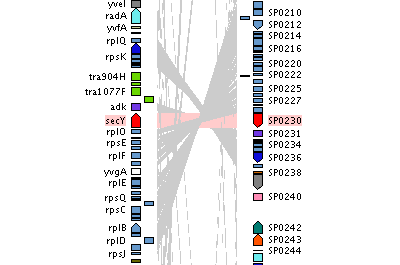
Comparison of bacterial genomes
Documentation
‘BugView’ has a brief set of internal Help menu items, with full documentation in the form an illustrated manual available as a pdf file that can either be downloaded or viewed on-line .
A short description of ‘BugView’ was published as an application note in Bioinformatics (2004) 20, 129-130. It was also discussed, with further illustrations of its use, in The Biochemist (2004) 26, 7-11, and described in more detail in Methods in Molecular Biology (2007) 395, 109–132. It was last tested successfully in 2023 (20 years after its initial release) using a GenBank file in contemporary format.
Downloads
‘BugView’ is available for the operating systems listed below. A run-time version of Java is required, and can be downloaded from Oracle.
- Mac OS X (Problems? Use the double-clickable Unix jar file)
- Unix/Linux
- Windows 32-bit
- Windows 64-bit
It helps to read the manual before using the program:
The Perl script, gcfprep, used in conjunction with standalone BLAST from NCBI, is also available.
Authorship and Acknowledgement
‘Bug View’ was developed by David P. Leader. It incorporates some code written by Andrei Grigoriev for the original ‘Der Browser’ Java applet.