Motivated Proteins
A Web Facility for Studying Small Hydrogen-Bonded Motifs

What motifs are these?
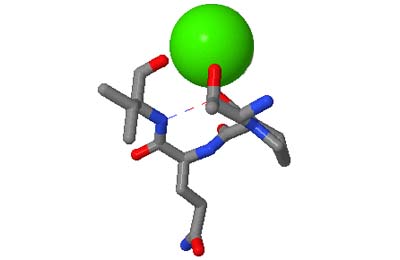
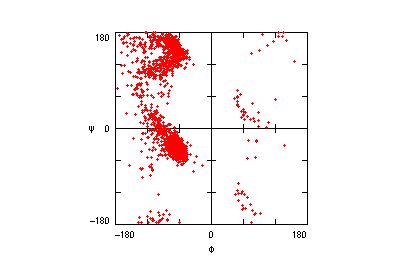
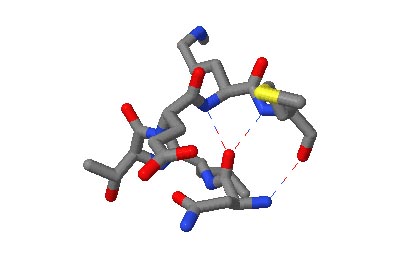
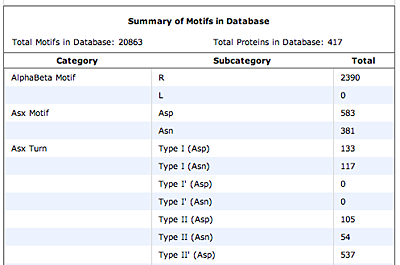
The motifs in question are small three-dimensional structures found in proteins. Typically they are loop-shaped, three to seven amino acid residues in length, and are generally stabilized by backbone — and in some cases side-chain — hydrogen bonds. More well-known examples include Beta Turns and Schellman Loops; others involving main-chain hydrogen-bonds include Beta Bulge Loops, and those involving side-chain hydrogen bonds include Turns, Loops and Motifs with Asx or Ser/Thr residues. In some motifs the three-dimensional structure can also be stabilized by linkages to partially charged atoms of ligands. Although some of the motifs were described some time ago, there are several interesting more recent additions.
A Motif Glossary with formal definitions and illustrations of each motif is available, and can be invoked in a pop-up window at any time when using the actual facility.
What do the facilities provide?
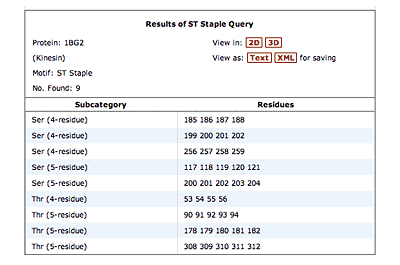
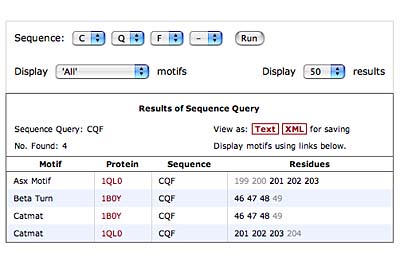
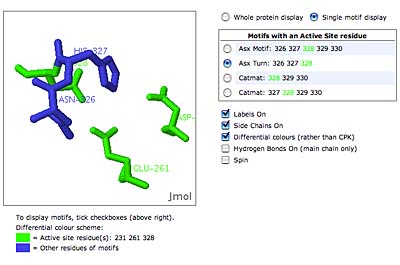
The original facility provides different ways of looking at the motifs in the context of a particular protein. You can list the motifs in a protein, see them in the context of the secondary or tertiary structure, and examine the three-dimensional structure of a specific motif and its hydrogen bonds in atomic detail. You can search for proteins with motifs near active sites or near ligands, search for proteins with motifs having a particular amino acid sequence pattern, or perform searches for overlapping motifs or motifs of one or the other main-chain enantiomeric forms. Summaries of amino acid compositions and dihedral angles are also available. A description of the facility has been published in BMC Bioinformatics (2009) 10:60.
The original facility has more recently (2021) been complemented by Motivated Proteins 2, which has a ten-fold larger database and integration with the SCOPe architectural classification of proteins. It adapts well to mobile devices, which the structure of original does not allow. It lacks a few features of the original that required manual curation or are best explored using our desktop application, Structure Motivator (also with an updated version).
Click on the thumbnails below for screen-shots illustrating features of the facility.
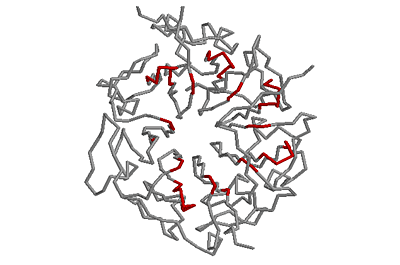
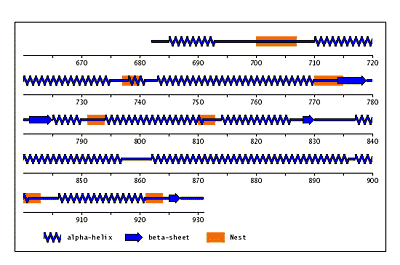
Beta-bulges in the 7-bladed beta-propeller of protein 1A12
Why the name?
We wished to spare users yet another acronym, but felt it best to avoid a name incorporating the term ‘motif’, which is often used for amino acid sequence patterns. We trust that the apparent frivolity of the name ‘Motivated Proteins’ is more than compensated for by its memorability.
Who provides these facilities?
David Leader and the late James Milner-White, of the College of Medical, Veterinary and Life Sciences in the University of Glasgow, created Motivated Proteins in 2008.